PHAT (Panchromatic Hubble Andromeda Treasury)

Description
The Panchromatic Hubble Andromeda Treasury is a Hubble Space Telescope Multi-cycle program to map roughly a third of M31's star forming disk, using 6 filters covering from the ultraviolet through the near infrared. With HST's resolution and sensitivity, the disk of M31 is resolved into more than 100 million stars, enabling a wide range of scientific endeavors.
The phat_v3.phot_mod and phat_v2.phot_mod tables have been crossmatched against our default reference datasets within a 1.5 arcsec radius, nearest neighbor only. These tables will appear with x1p5 in their name in our table browser. Example: phat_v3.x1p5__phot_mod__gaia_dr3__gaia_source.
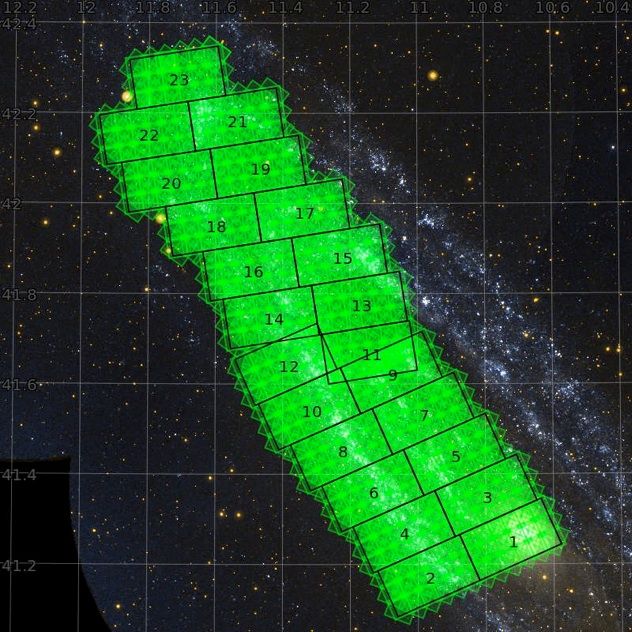
PHAT Brick coverage:
Scientific Goals
- Star formation histories derived on 50-100 parsec scales
- Improved stellar evolution models, calibrated at UV through NIR wavelengths
- Well-defined catalogs of stellar clusters, at all ages
- Characterization of variations in the stellar mass function from ∼3 to 30 solar masses
- Measurements of the mass function and age distributions of stellar clusters
- Maps of extinction from dust, and characterization of the extinction law
- Calibration of star formation indicators
- Age dating of supernova remnants
- Quantitative constraints on the coupling between star formation and the interstellar medium
- Identification and characterization of variable stars
- Kinematic decompositions of structural components
- Cross-identification of multi-wavelength sources and emission line objects
Data Releases
All PHAT data products are available as files from the MAST Archive. The Data Lab service provided here is database access to the photometric catalog, including the combined photometry for phat_v2 and phat_v3 and the single epoch measurements for phat_v2.
PHAT v3
Documentation for PHAT_v3 can be found in Williams, B.F. et al. 2023.
| PHAT_v3 Summary | |
|---|---|
| Area covered | 0.5 deg² |
| Bands | J0275W, J0336W (WFC3/UVIS), J0475W, J0814W (ACS/WFC), J0110W, J0160W (WFC3/IR) |
| Number of bricks | 23 |
| Number of objects | ~137,000,000 |
| PHAT_v3 Tables | |
|---|---|
| phot_mod | Combined average photometry |
PHAT v2
Documentation for PHAT_v2 can be found in Williams, B.F. et al. 2014.
| PHAT_v2 Summary | |
|---|---|
| Area covered | 0.5 deg² |
| Bands | F275W, F336W (WFC3/UVIS), F475W, F814W (ACS/WFC), F110W, F160W (WFC3/IR) |
| Depth (5σ, F275W, F336W, F475W, F814W, F110W, F160W) | 25.1, 24.8, 27.9, 27.1, 25.0, and 24.0 mag |
| Spatial resolution (F275W, F336W, F475W, F814W, F110W, F160W) | ∼0.08, 0.08, 0.1, 0.1, 0.25, and 0.25 arcsec |
| Number of bricks | 23 |
| Number of objects | ~117,000,000 |
| Number of measurements | ~7,500,000,000 |
| Photometric precision | ~1% |
| Astrometric accuracy | ~5 mas |
| PHAT_v2 Tables | |
|---|---|
| phot_meas | Individual photometric measurements, one row per measurement |
| phot_mod | Combined average photometry |
Data Reduction
The PHAT survey, the data reduction procedure, and photometric pipeline are described in the following papers:
Data Access
The PHAT data are accessible by a variety of means:
Data Lab Table Access Protocol (TAP) service
TAP provides a convenient access layer to the PHAT catalog database. TAP-aware clients (such as TOPCAT) can point to https://datalab.noirlab.edu/tap, select the phat_v3 or phat_v2 database, and see the database tables and descriptions. You can also view the PHAT tables and descriptions in the Data Lab table browser.
Data Lab Query Client
The Query Client is available as part of the Data Lab software distribution. The Query Client provides a Python API to Data Lab database services. These services include anonymous and authenticated access through synchronous or asynchronous queries of the catalog made directly to the database. Additional Data Lab services for registered users include personal database storage and storage through the Data Lab VOSpace.
The Query Client can be called from a Jupyter Notebook on the Data Lab Notebook server. Example notebooks are provided to users upon creation of their user account (register here), and are also available to browse on GitHub at https://github.com/astro-datalab/notebooks-latest.
Jupyter Notebook Server
The Data Lab Jupyter Notebook server (authenticated service) contains examples of how to access and visualize the PHAT catalog:
FTP Access
All of the PHAT catalog files, images, and products of the photometric pipeline are available as files on the MAST site.
Acknowledgments
Based on observations made with the NASA/ESA Hubble Space Telescope, obtained [from the Data Archive] at the Space Telescope Science Institute, which is operated by the Association of Universities for Research in Astronomy, Inc., under NASA contract NAS 5-26555. These observations are associated with the Panchromatic Hubble Andromeda Treasury Multi-cycle Program. Database access and other data services were provided by the Astro Data Lab. NOIRLab is operated by the Association of Universities for Research in Astronomy (AURA) under a cooperative agreement with the National Science Foundation.
